

Springer, Berlin, Heidelberg, pp 130–137Īnbarasu LA, Narayanasamy P, Sundararajan V (2000) Multiple molecular sequence alignment by island parallel genetic algorithm. In: McKay B, Yao X, Newton CS, Kim J-H, Furuhashi T (eds) Simulated Evolution and Learning. Īnbarasu LA, Narayanasamy P, Sundararajan V (1999) Multiple sequence alignment using parallel genetic algorithms. IEEE Trans Paral Distrib Syst 9(3):283–294. Yap TK, Frieder O, Martino RL (1998) Parallel computation in biological sequence analysis. In: Hertzberger B, Sloot P (eds) High-Performance Computing and Networking. Martino RL, Yap TK, Suh EB (1997) Parallel algorithms in molecular biology. Hughey R, Krogh A (1996) Hidden Markov models for sequence analysis: extension and analysis of the basic method. In: Proceedings of the 1995 International Conference on Parallel Processing ICPP Yap TK, Munson PJ, Frieder O, Martino RL (1995) Parallel multiple sequence alignment using speculative computation. Ishikawa M, Toya T, Hoshida M, Nitta K, Ogiwara A, Kanehisa M (1993) Multiple sequence alignment by parallel simulated annealing. ĭate S, Kulkarni R, Kulkarni B, Kulkarni-Kale U, Kolaskar AS (1993) Multiple alignment of sequences on parallel computers. Tajima K (1988) Multiple DNA and protein sequence alignment on a workstation and a supercomputer.
#Multiple sequence alignment software#
Special Section: Component-Based Software Engineering (CBSE), 2011īornmann L, Daniel H-D (2007) What do we know about the h index? J Am Soc Inf Sci Technol 58(9):1381–1385. Mahdavi-Hezavehi S, Galster M, Avgeriou P (2013) Variability in quality attributes of service-based software systems: a systematic literature review. įlores-Contreras J, Duran-Limon HA, Chavoya A, Almanza-Ruiz SH (2021) Performance prediction of parallel applications: a systematic literature review.
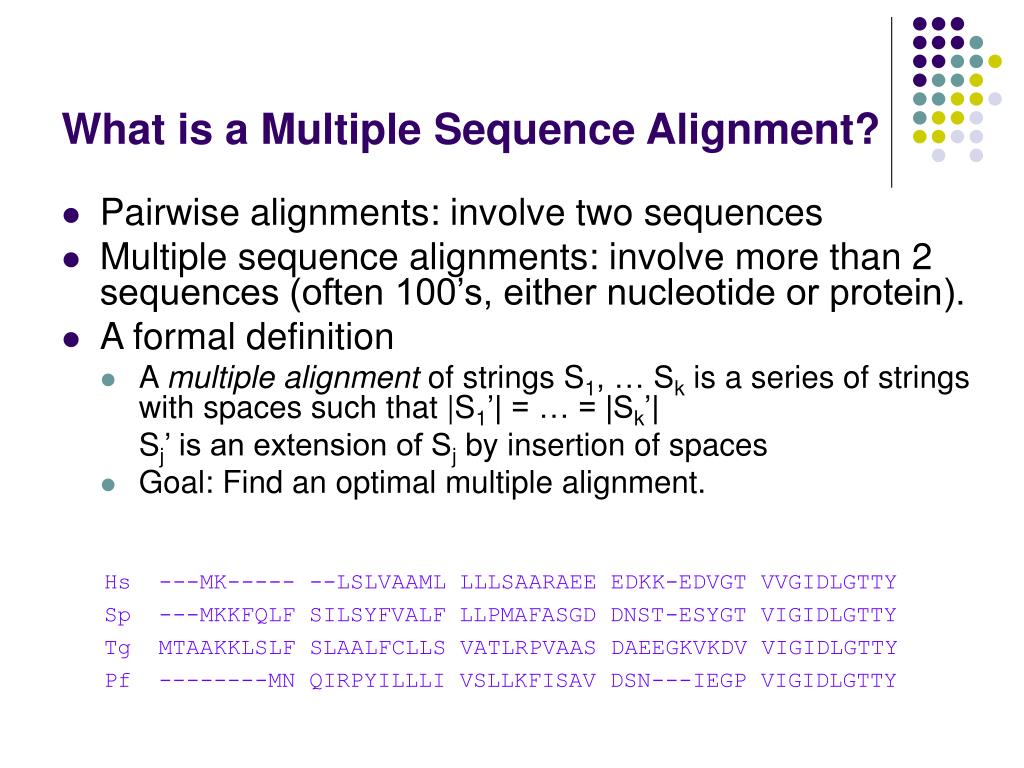
Hujainah F, Bakar RBA, Abdulgabber MA, Zamli KZ (2018) Software requirements prioritisation: a systematic literature review on significance, stakeholders, techniques and challenges. ĭe Freitas Junior M, Fantinato M, Sun V (2015) Improvements to the function point analysis method: a systematic literature review. Galster M, Weyns D, Tofan D, Michalik B, Avgeriou P (2014) Variability in software systems–a systematic literature review. Hall T, Beecham S, Bowes D, Gray D, Counsell S (2012) A systematic literature review on fault prediction performance in software engineering. Salleh N, Mendes E, Grundy J (2011) Empirical studies of pair programming for CS/SE teaching in higher education: a systematic literature review. Special Section: Software Engineering track of the 24th Annual Symposium on Applied Computing Ĭhen L, Ali Babar M (2011) A systematic review of evaluation of variability management approaches in software product lines. Technical Report EBSE 2007-001, Keele University and Durham University Joint Report. Kitchenham BA, Charters S (2007) Guidelines for performing systematic literature reviews in software engineering. Ībascal F, Zardoya R, Telford MJ (2010) TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Wernersson R, Pedersen AG (2003) RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. īonizzoni P, Vedova GD (2001) The complexity of multiple sequence alignment with SP-score that is a metric. Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Cambridge University Press, Cambridge, England. ĭurbin R, Eddy SR, Krogh A, Mitchison G (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Ramsden J (2009) Bioinformatics: An Introduction, 2nd edn. Additionally, we point out some directions and trends for parallel computing approaches for multiple sequence alignment, as well as some unsolved problems.īayat A (2002) Bioinformatics. We conducted a selection process that yielded 106 research articles then, we analyzed these articles and defined a classification framework. Additionally, in order to cover other potential databases and journals, we performed a transversal search through Google Scholar. In this paper, we present a systematic literature review of parallel computing approaches applied to multiple sequence alignment algorithms for proteins, published in the open literature from 1988 to 2022 we extracted articles from four scientific databases: ACM Digital Library, IEEE Xplore, Science Direct and SpringerLink, and four journals: Bioinformatics, PLOS Computational Biology, PLOS ONE, and Scientific Reports. Since multiple sequence alignment has exponential time complexity when a dynamic programming approach is applied, a substantial number of parallel computing approaches have been implemented in the last two decades to improve their performance. Multiple sequence alignment approaches refer to algorithmic solutions for the alignment of biological sequences.


 0 kommentar(er)
0 kommentar(er)
